SIRT3/Ac-CS2/NAD+/isoNAM is also provided. Binding affinity_SIRT3 with small inhibitor.docx
XG(3/25/14):Protocol of isoNAM-SIRT1 relative activity. Protocol of isoNAM-SIRT1 relative inhibition.docx
PL(03/25/14): Report on structural alignment and C pocket analysis. C pocket.docx
RC (3/25): A few questions:
a) for the free SIRT3/ternary SIRT3 comparison, do the residue contacts to NAM change due to movement of the loop? You might consider scoring a few molecules from the current list of top hits for SIRT3 against the free SIRT3 structure to see the effect of the movement of the loop on screening (e.g., to see if the rank ordering changes)
PL(03/26/14): I did use different structures as receptor for docking and MM-GBSA calculations before using Schrodinger's programs. Results are attached.
docking results for different SIRT3 structures.docx. The ranking usually did not change, probably due to error cancellation. The relative differences, however, do change.
b) for the 4FVT/1YC2 A comparison, what is being shown? Is it primarily the difference between nicotinamide in NAD+ vs free NAM? Does chain A contain PEG?
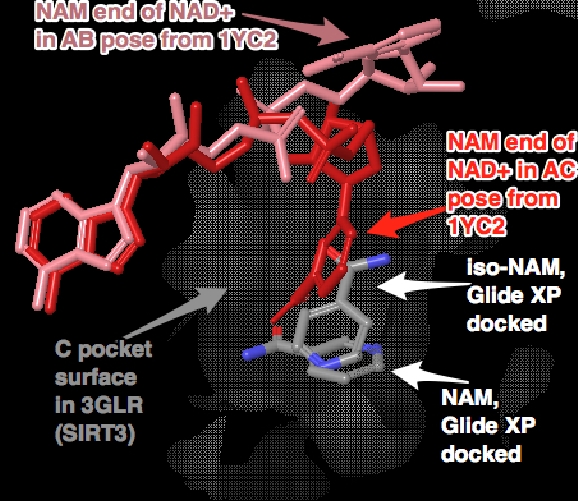
PL(03/26/14): The 4FVT/1YC2:A comparison shows the changes of nicotinamide positions. Free nicotinamide (as in 1YC2:A) goes lower into the pocket. The interaction diagrams [as in 2) and 3)] also differ in surrounding residues. In 1YC2:A, there is no long PEG chains, but do have ethylene glycol in place.
c) for the SIRT1/SIRT1:ligand complex comparison, can the changes in the C pocket be analyzed further?
PL(03/26/14): can you be more specific about the expected analysis? As you can see, using Rossmann fold domain for alignment, the main difference in structure is in the relative position of the whole Zn-binding domain. The impact of the C pocket is mainly on the loop region around the binding site, starting from the hinge loop. Also, NAD+ seems to bind stronger to the SIRT1 than SIRT3, as it does not require extra PEG or Glycol in the place of peptide binding site to help the protein fold into the close form.
Related: for the C pocket binding ligands you have screened so far, can you comment on the differences between SIRT3 MM-GBSA scores for enzyme vs enzyme-intermediate complexes? Specifically, have you found any ligands that display a significant difference in these scores, preferentially binding to the enzyme-intermediate complex?
PL(03/26/14): Almost none. There is only one molecule shown below, that displays preferred interaction with enzyme-intermediate complex in its positively charged form. Here the docking and MM-GBSA calculations are carried out using 4BVG structure, with or without intermediate.
 |
| (1-Isobutylpiperidin-3-yl)methanol |
RC (1-14): Plan updates based on meeting.
Experimental -
-- Based on information regarding ITC operating range, it is important that we determine whether ITC can measure NAD binding affinity to SIRT1 in absence of peptide as soon as possible, given
that data suggests that NAD-SIRT1 is the tightest binding complex (in presence of peptide) that we are currently looking at. Since the experiment may not work, we need to leave time for other experiments (below).
-- Since peptide binding affinity to SIRT3 has already been determined by ITC, we will not consider it at this time
-- If the above is not possible, we will move on to inhibition kinetic studies with isoNAM/SIRT3. Please let me know whether it would take a month for just SIRT3 or whether you meant 1 month for both SIRT1 and SIRT3.
-- Prior to doing inhibition kinetics studies, we should finish the relative inhibition experiments with isoNAM.
-- Please provide a schedule for the above studies so I can determine whether they will fit within our allotted time. If not, we may rearrange tasks.
Computational -
-- We have already run MD binding affinity calculations on SIRT3/peptide/NAD/NAM. Based on the fact that ITC cannot accurately measure binding affinities for NAM/isoNAM, and the fact that there are no new contacts observed between intermediate and NAM/isoNAM, if XG confirms that it is not possible to directly measure mM binding affinities experimentally with other techniques, there is no particular reason to start binding affinity calculations on the intermediate complexes at this time. Therefore we should dedicate as much GPU time as possible to the SIRT3/peptide/NAD/isoNAM complex to start.
-- Regarding isoNAM as a base exchange substrate, please post the paper by Denu on base exchange substrates or point out where it is on the wiki.
-- Given that it is likely not possible to determine NAD binding affinity to SIRT3 in absence of peptide with ITC, I would like to consider whether it will be possible to run an MD calculation on NAD in AC pocket in absence of peptide, for comparison to the ternary complex already studied. The aim of this study would be to provide some explanation from a computational standpoint as to why the binding affinity changes substantially in absence of peptide. Please let me know if we have the required apo crystal structure and if we can prepare an appropriate starting structure for this calculation.
-- If the latter is not possible, the next MD calculations to do after the SIRT3/peptide/NAD/isoNAM complex should be NAM/isoNAM with 1YC2.
-- Similarly, please provide schedule of work so we can determine how the computational schedule will align with that for experiments.
-- Protocols for the MD binding affinity calculations should be written in a format that can be used in the paper.
Please prepare a new wiki page so we can separate the correspondence on this paper from that for "project 1".
RC (1-6):
I am planning to ask for an extension on the resubmission of the sirtuin paper.
Our proposed mechanism was based on some of the earlier literature from the Sinclair group which has now been rendered obsolete, but which was not explicitly refuted in later work on Sir2/SIRT1 inhibition mechanisms, leading to some confusion in the field. Also, as noted earlier on the wiki, some critical details of the structural literature were not carefully read at the time when the docking studies were set up - in particular, the fact that peptide binding forces NAD into the AC pocket was not made clear when our computational studies were initially planned. This can perhaps be attributed to the work involved in entering a new field and launching both computational and experimental investigations of a new enzyme simultaneously.
I have only recently had time to carefully read the literature myself. The details of binding affinities of NAM and NAD to the various pockets, as well as the values of exchange equilibrium constants, play an important role in the inhibition mechanism for NAM. Though many of these details are not currently known for SIRT3, the data available for Sir2, as well as the isoNAM data we collected for SIRT3, suggest that binding affinities of NAM and isoNAM are low, and that base exchange plays an important role in observed inhibition kinetics for NAM. A simple picture of competitive inhibition, through competition with the substrate for the active site, may apply to SIRT3 as well, but for inhibitors other than NAM. Our initial intent was to study multiple inhibitors in order to isolate these effects, but we did not complete those experiments, given the time required to help Eric finish his part of the paper.
The noncompetitive inhibition by NAM discussed in prior literature is based on standard models for noncompetitive inhibition, or an informal use of the term noncompetitive. This has contributed to the confusion in the field regarding sirtuin inhibition mechanisms. Reading of the literature indicates that our work has provided an opportunity to clarify some of the confusion regarding sirtuin inhibitory mechanisms through a more careful analysis of SIRT3 inhibition. Also, as you know, there is overlap between aspects of the kinetic modeling we were planning to put in the 2nd paper and the inhibition mechanism discussed in this paper. (More generally, this is part of our group's overall interest in MM experiments, parameter estimation and kinetic modeling of other enzymes with complex mechanisms, like the polymerases in Chaoran's extension kinetics project.) We need to plan out future work in a manner consistent with the material presented in this paper, and correcting some of the above issues will be easier now than later. Due to the significant, growing interest in this area, and our intention to model activation, it is important that our early work is consistent with the work that we plan to do next.
We can make inferences about SIRT3 inhibition mechanisms based on our current data and literature data on Sir2/SIRT3, but in the absence of more direct measurements and calculations, our scientific contributions in this paper are not clear and are not quite up to the standards of prior mechanistic work on Sir2. However, given our work to date, we are now in a position to rapidly collect both targeted computational and experimental data that will go a long way toward making a more compelling case for a particular inhibition mechanism in SIRT3. We should take this opportunity to report some of these measurements and computations that we have been planning before someone else does, given their important implications for SIRT3 activity modulation.
These measurements/calculations may include MD computations of binding affinities of NAM/isoNAM, ITC measurements of the latter, isoNAM relative inhibition experiments, and isoNAM inhibition mode experiments. isoNAM provides an ideal point of comparison for mechanism identification, since binding modes may be quite similar, despite the substantial differences in inhibitory potency. Please start on the simulations and experiments requested on the wiki and provide an expected times for the completion of each, since these times are relevant to the extension request. Please also reply to all the questions I have posted there. Newly requested figures and methods write ups are still relevant and required. In the meantime I will revise the paper to render it consistent with the literature and clarify some of the misunderstandings and oversimplifications of inhibition mechanisms in the prior literature. I will also provide more commentary on how the new data will be used in the paper. This will set the stage for a more complete kinetic modeling in the next paper (vis-a-vis activation).
Once we have some new results and estimates of the time required for the above measurements and simulations, I will provide a more detailed schedule for experiments and computations including a rank ordering by their priority, since not all of this data will be included in this paper. At that point we may start a new wiki page dedicated exclusively to the revisions of this paper.
My extension request will focus on the time required to complete MD simulations requested by referee 1 (to be conservative, I am requesting a 2 month extension).
Raj
RC (1-9): XG, I would like to make a list of the possible ITC experiments that we can do (in principle). Is the following correct (each applies to both SIRT1 and SIRT3):
E-NAM/isoNAM: possible
E-NAD: possible
E-peptide: possible
E-peptide-intermediate-NAM/isoNAM: not possible since intermediate cannot be trapped
E-peptide-NAD: not possible since reactive complex
XG (1-09) Thank you for the detailed list.
(1) To confirm: E-NAM/isoNAM = E-NAM and E-isoNAM.
(2) E-peptide-NAD can be indirectly measured by using Carba-NAD.
RC (1-9): Availability of Carba-NAD.
XG(1-13): Carba-NAD+ consists of a NAD+ molecule where the oxygen atom of the nucleotide has been replaced with a carbon atom.The substitution prevents the hydrolysis of nicotinamide and the ribose mimic.
Carba-NAD+ is not commercially available. The synthesis methods can be found in the following references:
(1) Slama JT, Simmons AM. Carba-NAD: synthesis and enzymological properties of a carbocyclic analogue of oxidized NAD. Biochemistry 1988 27(1): 183-93.
(2) Szczepankiewicz BG et al. Synthesis of Carba-NAD and the structures of its ternary complexes with SIRT3 and SIRT5. Journal of Organic Chemistry, 2012, 77, 7319-7329.
Few researches included Carba-NAD+.
(1) In Landry 2000 Biochem. BioPhys Res.Com Paper, Carba-NAD+ was tested as a yHst2 inhibitor and was discovered to compete for the coenzyme binding site within the active site.
(2) In Zhao’s 2004 PNAS paper, crystal structure of the Saccharomyces cerevisiae Sirt2 homologue, yHst2, bound to an acetyllysine-containing histone H4 peptide and carbaNAD+ was reported. The dissociation constant Kd of the reactions (a. ADP-Ribose with,yHst2. Kd=0.404 uM; b. ADP-Ribose with binary yHst2/k16-acetylated histone H4 peptide. Kd=29.16 uM) were reported by using VP-ITC.
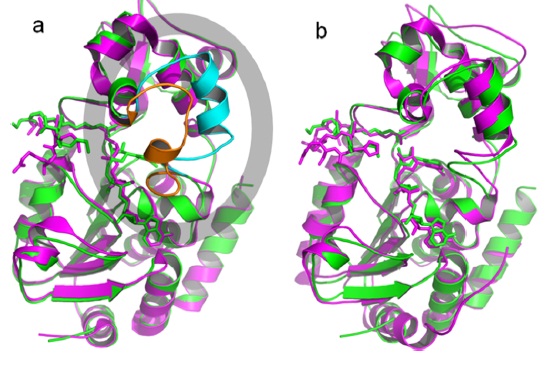
(3) In Szczepankiewicz 2012 J. Org Chem. Paper, the Xray crystal structures of the ternary complexes of SIRT3 (4FVT) and SIRT5 (4G1C) bound to a peptide substrate and carba-NAD were reported. Structural comparison of the SIRT3/ACS2/carba-NAD ternary complex. (a) Comparison of the structure of SIRT3/ACS2/ carba-NAD (green/cyan) with that of SIRT3/imidate intermediate (magenta/orange). The cyan-colored helix in the gray oval uncoils to render the orange loop after nicotinamide displacement. (b) Comparison of the structure of SIRT3/ACS2/carba-NAD (green) with that of yHst2/H4/carba-NAD (magenta).
RC (1-15): It should be possible to have a CRO synthesize carba-NAD for us, though not for this paper. There are a lot of organizations doing custom synthesis these days. If we can confirm that ITC can measure in the low hundreds of uM, we should look into this when we have time (not a priority right now, but let's not forget in case it's possible), given the relevance of these numbers for sirtuin mechanism and regulation.
Have any of the above ITC measurements been reported in the literature on SIRT1 or SIRT3?
XG (1-14): ITC experiments overview
Inhibitor studies:
*In Cosgrove Biochemistry 2006 paper, the binding constants of different peptide substrate with Sir2Tm wild-type and mutant proteins were measured using VP-ITC.
*In Jin et al JBC 2009 paper (Crystal structures of human sirt3 dispalying substrate-induced conformational changes, JBC 284: 24394-24405), substrate binding study by ITC was reported. The ITC experiments were performed by titrating the AceCS2-K ac peptide or NAD+ to SIRT3. The peptide binds to SIRT3 with binding constant of 15 uM. NAD did not bind to SIRT3 directly. Please check the attached paper for detail it was on page 24402.
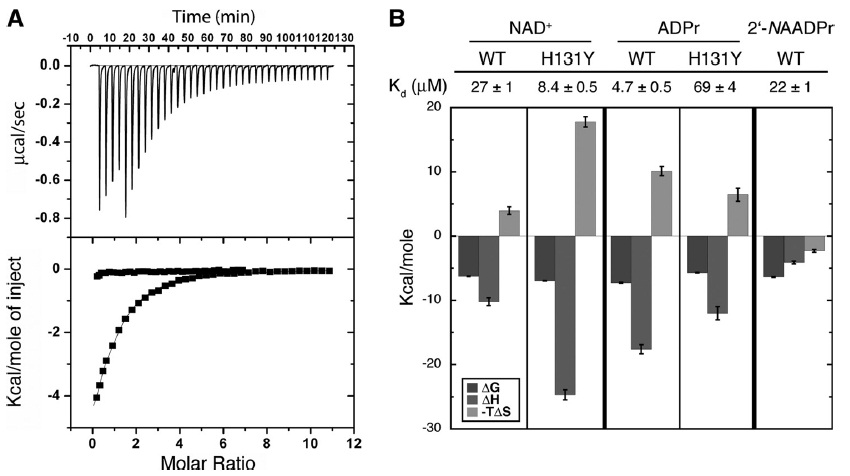
*In Pan PW JBC 2011 paper, 5 binding pairs (NAD- WT SIRT6, NAD-Hst2, 2’-NAADPr – WT SIRT6, NAD-H131Y SIRT6, ADPr-H131Y SIRT6) were investigated. NAD+bound to SIRT6 with a Kd of 27 ±1 mM (Fig.5B), providing the first physical evidence that SIRT6, unlike other characterized sirtuins, can bind NAD+ efficiently in the absence of an acetylated substrate. Figure below: The change in Gibbs free energy (DG) is shown in black. Change in enthalpy (DH) is shown is gray, and-TDS is displayed in light gray.
*In Ex-527 PNAS 2013 paper, the binding experiments of Ex-527 with SIRT3 and Sir2Tm (not E-NAD) were done by using MST. Ex-527 showed negligible affinities (Sir2Tm: Kd > 180 μM; Sirt3 Kd > 330 μM) to apo-enzymes and to the Sirtuins saturated with 1 mM peptide substrate (Sir2Tm: Kd > 170 μM; Sirt3: Kd > 180 μM). Adding saturating NAD+ concentrations (5 mM), however, improved Ex-527 binding, with Kd values of 6.0 ± 0.4 μM for Sir2Tm and 16.5 ± 2.9 μM for Sirt3. Strikingly, addition of both substrates resulted in even higher affinities, with Kd values of 4.9 ± 0.5 μM (Sir2Tm) and 10.0 ± 1.4 μM (Sirt3).
Activator studies
*In Sritris 2007 Nature paper (Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes, 450: 712-716), binding constant of SIRT1-C–peptide substrate complex with the activator SRT1460 was investigated using VP-ITC. The titration of SRT1460 against purified human SIRT1-C did not result in detectable binding. In the presence of acetylated peptide substrate SRT1460 exhibited signs of protein-binding-site saturation best fitting a one site binding model. The dissociation constant (Kd) for SRT1460 was 16.2 mM and the enthalpy (DH) was26.1 kcal mol-1.
*In Sinclair 2013 Science paper (Evidence for a Common Mechanism of SIRT1 Regulation by Allosteric Activators, 339: 1216-1219), Isothermal titration calorimetry (ITC) did not detect binding between saturating amounts of PGC-1a peptide (2 mM) and STAC-1 (100 mM) or STAC-2 (50 mM.
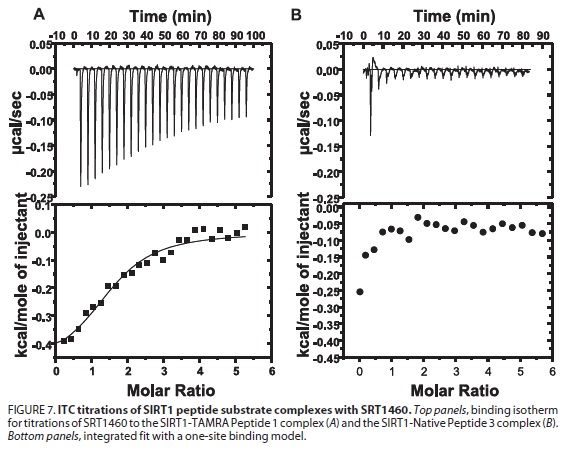
*In Pfizer 2010 JBC paper (SIRT1 Activators Do Not Stimulate Activity with Native Substrates 285: 8340-8351), ITC was used to define the binding energetics of compounds to the SIRT1 peptide substrate complex. ITC Studies Demonstrate That SRT1460 Binds to the SIRT1-TAMRA Peptide Substrate Complex but Not to the SIRT1-Native Peptide Substrate Complex. The ITC study demonstrated that the binding of SRT1460 is observed only in the presence of the fluorophore-containing peptide substrate.
Summary:
(1) VP-ITC has been used in Sirtuin field and data were reported on good journal like Nature, Science, PNAS …
(2) The applications of VP-ITC include (a) small molecule-enzyme, (b) substrate-enzyme.
(3) From the reported data, it tells that the upper bound of VP-ITC sensitivity is ~ 100uM. There was very few to report the exact number beyond that range.
What ITC experiments we can do is relevant to the decision of what MD binding affinity calculations to do.
Please also provide an ITC protocol.
XG (1-09): Start up protocol is attached. ITC protocol.pdf
RC: Please also review the ITC operating range (which we discussed once previously) vis a vis the above experiments.
We should have the ITC updates above (and I would like to review them) prior to meeting since they are relevant to the plan.
XG (1-13):
Operating range of VP-ITC: 2-80oC for temperature.
Sensitivity of VP-ITC: For new machine, it was claimed that it can directly measures millimolar to picomolar binding constants. However, I do prefer, as Chaoran mentioned, the comfortable zone for accurate measurement will be 1nM - 100uM (or up to 200 uM).
From: Chaoran Jing
Sent: Fri 12/27/2013 4:50 PM
To: Raj Chakrabarti
Hi, Raj
Based on my experience, it usually takes a couple of hours to run one sample, including system cleaning. For each binding pair, if everything goes smoothly, one would need at least two samples: one with small molecule, one reference without small molecule. One may also need to adjust concentration of protein and/or small molecules for best S/N, which would take more time. Typically people plan for at least one day experiment for each binding pair.
My impression is that ITC works well from low nM to low uM. It can measure binding affinity up to hundreds of uM or even low mM. However, at this scale the non-specific binding may dominate. Also the higher the Kd, the higher the concentration needed for ITC sample. Usually the protein concentration obtained from small scale protein prep is ~ 10-200uM. Some protein may crash out at high concentration. These factors limit the detection limit to about hundreds of uM. For reliable results, I would say, 1nM - 100uM.
Regards
Chaoran
RC (1-13): There are still some experimental issues that XG is looking into, which are relevant to our planning. The meeting time will be set after I receive that information. In the meantime, MD simulation preparation, figure revision, software (windows matlab) installation, and any pending docking studies requested should proceed.
RC (12-27):
I am working on making paper revisions that render the presentation/interpretation of our results consistent with the literature, and need some related data.
1) Computational results:
n Have we recorded the MM-GBSA scores for NAM and isoNAM in Sir2, SIRT3 C pocket in the presence of ADPR intermediate in A pocket? Have seen the scores in the absence of intermediate. Any evidence from docking that NAM or isoNAM engage in additional stabilizing interactions w ADPR?
PL(12/27): I do not have MM-GBSA scores for NAM and isoNAM in Sir2, and do have these scores in SIRT3 C pocket with or without the presence of Intermediate (from structure 4BVG):
SIRT3(4BVG) with intermediate SIRT3(4BVG) as apo enzyme
NAM: -38.55 -39.81
isoNAM: -32.36 -34.68
There is no calculation done for intermediate's binding, therefore there is no information on their stabilization effect on intermediate.
n Related: recall that there was flexibility in the binding modes of NAM, isoNAM to enzyme in the absence of intermediate; i.e., there were at least two docked poses with significant RMSD but similar scores. Is this also the case for ADPR intermediate complex w these molecules, or do you find a single pose?
PL(12/27): For SIRT3(4BVG) with intermediate (Alkylimidate), there are two poses different significantly in orientations with similar scores in both cases of NAM and isoNAM.
NAM (pose 1): -38.55
NAM (pose 2): -36.74
isoNAM (pose 1): -32.36
isoNAM (pose 2): -28.17
RC (12-27): Do these two conformations correspond to the "flipped" conformations shown by Wolberger on pg 864 of paper posted below? If so do both isoNAM and NAM adopt the flipped conformational stabilized by additional interaction involving the ring nitrogen?
PL(01/07/2014): No, the two conformations do not correspond to the flipping proposed in the scheme on page 864. The two conformations observed in docking involve the shift of amide group instead of simple fliping of the ring.
RC (12-31): In Wolberger 2005, p 865, there is a proposal regarding "contracted" and "extended" conformations of the intermediate. The extended form, which was reconstructed based on positions of the NAD and acetyl lysine in crystal structures, features the ADPR and NAM in closer proximity suitable for reaction. It appears the extended form was not observed crystallographically, but it may be relevant to the analysis of the binding of NAM/isoNAM to the intermediate discussed above. One may assess whether the ADPR in SIRT3 structures above is closer to the contracted or extended forms in Sir2.
RC (1-9): I suggested the analysis above in order to determine whether the extended conformation of the intermediate may confer some stabilization of NAM/isoNAM binding to the intermediate complex, since the docking/MM-GBSA results so far did not seem to indicate any new contacts between these inhibitors and the intermediate complex that were not already present in the EI complex. This is relevant to the priority of running MD simulations on the intermediate complex as well as the EI complex.
RC (12/27): Toward the goal of assessing the differences between NAM/isoNAM binding as they affect the mechanism of inhibition, please describe the similarities/differences in protein-ligand interactions for NAD/isoNAM in the absence of intermediate (I believe this was in an earlier draft prepared by Eric, but may not be in the current one). Please prepare a figure that shows the interactions for each, if Eric did not already prepare one. We may proceed to set up MD simulations to obtain binding affinity estimates for isoNAM in the C pocket in the absence of ADPR, first for SIRT3 and then for Sir2. (I believe we already have MD binding affinity estimates for NAM/SIRT3.) These are closely related to the MD simulations we are planning to run on C pocket inhibitor leads. Please comment on how long it will take to set up and run a MM-PB(GB)SA calculation for isoNAM/SIRT3 binding affinity calculation. Please also update regarding the status of the GPU node and whether we can start using it for MD. We may consider running several MD simulations in parallel. In the meantime Autodock or DOCK docking of these compounds should also be set up for comparison to Glide as we discussed.
PL(01/07/2014): For NAM/isoNAM interactions with proteins, do you expect an interaction map from docking or from known crystal structures? Here is one figure I from from Eric's work.
RC: The above is probably sufficient for now.
For MD simulations, do you want to set up simulations with Sir2 as well? Our experimetnal results is on SIRT1, and the referenced structures (monomers from 1YC2) are from Sir2Af2, and some of the structures in the references below are for Sir2TM. We could do some simulations with SIRT1 since there are more structures available now, although no ternary crystal srtucture available makes it more difficult to obtain highly accurate complex structure.
RC (1/9): We should confirm that there is experimental inhibition kinetics data in the literature for the Sir2 isoforms above (in this case, given that our existing computational studies were done with Sir2, comparison to Sir2 binding affinities may be sufficient). Regarding SIRT1, please identify the structures we could use from the spreadsheet XG has prepared. If we do ITC measurements on SIRT1, binding affinity computations with that enzyme as well may be desirable. SIRT3 is higher priority, but as noted above we may do the same simulations on Sir2/SIRT1 if there is time. Once we have settled on the binding affinity calculation protocol we will decide when to do calculations on Sir2.
Yes, we have some MD binding affinity estimate for NAM/SIRT3 with the presence of acetylated peptide and NAD+.
For binding affinity estimates for NAM/isoNAM, we need to decide exactly what will be the receptor, w/ or w/o acetylated peptide, NAD+, intermediate, or even prodcuts (Ex527 binds better with the presence of product and forms a stable complex that block the continuation of reaction.)
RC (1/9): The reason I suggested binding affinity calculations on NAM/isoNAM in the absence of intermediate is that it will shed light on the role that direct competition of the inhibitors with NAD plays in the inhibition mechanism, when considered alongside the differences in experimental inhibition data for the molecules. These molecules are members of a congeneric series binding in the same pocket, making comparisons of their computationally estimated binding affinities more accurate. With acetylated peptide is preferred. Binding affinity in the presence of intermediate is also useful to estimate, since it is relevant to the base exchange mechanism. Based on the responses above, it seems that docking has not identified any contacts between the inhibitors and the intermediate (see above). So the calculations on the two complexes may be somewhat redundant. If we can do ITC measurements on the EI complex (in absence of intermediate), it may be preferred to MD calculations on the intermediate complex, since measurements cannot be made on that complex.
I had some limited results from Autodock, and I will try to make some comparison with Glide. However, I believe this is not our top priority right now.
PL(12/27): AADPR is the product, the intermediate I refer to is Alkylimidate. What is the ADPR that you referring here?
RC: Yes I'm referring to the same intermediate, with reference to the ADPR moiety (without NAM) in order to distinguish it from NAD.
Setting up MD simulations for isoNAM and the corresponding MM-PB(GB)SA calculation will not fit to our time frame for submission. The preparation of isoNAM force field parameter requires quantum mechanics calculation and charge fitting, will take a couple of days before actual simulation.
RC (12/31): Ok, please give me an idea of how long this calculation would take, as well as how long each additional binding affinity calculation of NAD in various pockets will take, leveraging the use of the new hardware, for the purpose of planning the next phases of the MD work.
PL(12/31): The force field parameterization for intermediate and isoNAM will take about one week. The intermediate will be more time-consuming the molecule is larger and many factors need to be considered.
RC (1/9): Please provide some more details regarding the factors mentioned. We should carry out force field parameterization of intermediate.
There are several factors affecting binding affinity calculation. If we are doing the same way Wolfgang Sippl's group have done for thiobarbiturates's binding to SIRT2, the MD (assume two ns) and MM-PB(GB)SA calculations for each molecule can be done in two days. If docking is required, it will take extra time.
RC (1/9): Yes, we should settle on an MD binding affinity calculation protocol. Please provide the protocol you used so far for binding affinity calculations on AcLys-peptide/NAD/NAM complex, if you haven't already provided it. Other than the duration of the simulation, how does your calculation differ from that of the Sippl group?
The priorities are isoNAM and NAM, for which we already have docked structures. After deciding on the protocol we will be able to schedule the MD binding affinity calculations in order of priority and decide whether we will have time to do calculations on NAD as well.
Currently there are some issues running NAMD on our system using GPU, some minimization and MD works fine, and using one GPU is about three times faster than using 12 CPUs on original nodes. However, there are some unexpected failures for some MD simulations. I have contacted the Exxact company for supports.
The GPU node is up and running now after I installed the CentOS 6 and CUDA 5.5. The process was delayed a bit by a bug in the NVIDIA driver shipped with CUDA tool kit. NAMD was recompiled to run on GPU. Currently, benchmark testing run is going on.
RC (1/9): Ok, pleease let me know the results of the test run when complete. Is it advisable to run only one MD simulation on the GPU node at a time; i.e., will our various MD simulations be run in series?
PL(01/09/14): There are still some jobs that runs on CPUs but not successful on GPUs. I do not have much time to investigate the problem at this moment. For those simulations that do run on GPU, the ENERGIES are comparable with those with CPUs, and I need to compare the structures and see if there is anything out of ordinary. MD simulations can run on either one GPU or both GPUs, and there is significant gain in using second GPU. In NAMD, you can specify which GPU card to use in the command, therefore, it is quite possible we can run two jobs at the same time on different GPUs, though I have not tested the impact to the wall time.
I just set up autodock software on PC, will begin some test runs next week.
n Please provide brief commentary regarding the robustness of the MM-GBSA scores to the choice of monomer, referring to the revised docking scores on the Ping’s tasks page.
PL(12/27): For complex with known crystal structure (even for cases using ligand analogs), the MM-GBSA scores based on crystal structures are found to correlate reasonably well with their binding affinities. (there is a reference can be sited here.) Our in-place MM-GBSA scores shows that there is little difference in using B or C chains as receptors for AC docking mode. In the case of AB docking mode, the nicotimamide moiety of NAD+ has two different orientations, and the one in chain A has a lower MM-GBSA score, and is best used to represent the binding affinity of AB binding mode.
n Please provide more info on the “AX” binding mode of NAD in SIRT3 and how it was prepared. From the wiki postings, I gather it was found in both docking and MD simulations. However, a Glide XP score was provided for the AX mode whereas a Glide SP score was provided for the other binding modes. Ideally, a figure with caption should be prepared highlighting the similarities/differences with respect to the AB binding mode (docked pose or the snapshot shown on the Project 1 page) in Sir2.
PL (12/27): The "AX" binding in docking: All reported values are all MM-GBSA scores (Glide SP or XP was used to generate poses). No Glide scores were reported. The receptor included SIRT3 and acetylated peptide from 4FVT, plus NAM docked to the C pocket (obtained from glide SP docking of NAM). The docking of NAD+ is performed using Glide SP. And the selected pose rank top in MM-GBSA score for NAD+ in its current form.
The "AX" binding in MD: the initial structure was constructed by superimposing NAD+ and NAM in AB binding mode (from 1YC2 chain A) into SIRT3 with peptide (from 4FVT), followed by MD simulation.
RC (12/27): Please provide a figure with a caption that depicts the AX binding mode and compares it to the AB binding mode. Where is the X pocket located, with respect to 'B pocket", C pocket, and peptide binding pocket? If docking of NAD+ was performed using Glide SP, why is there a reference to Glide XP in the reporting of the AX binding score? Please comment on where XP was used and why. I believe you mean NAD+ binding to the AX pocket is the top ranked pose in the presence of NAM (but is not observed in Glide results in the absence of NAM). Does binding to the AX pocket occur in absence of peptide, as does AB pocket binding in Sir2?
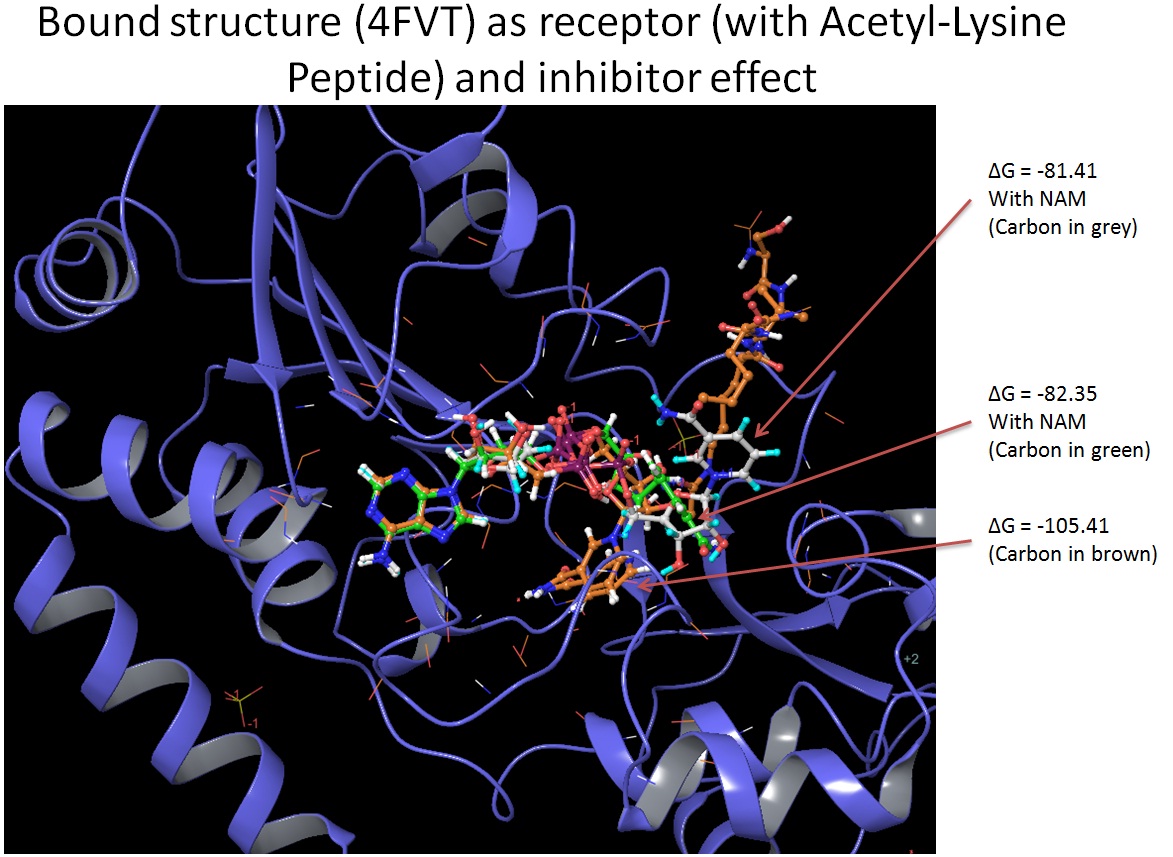
PL(12/27): Attached is the manual alignment between 1YC2 chain A (in brown color) (NAD+ in AB pocket, NAM in C pocket; green) and 4FVT (in colored based on secondary structure) with NAD+ docked into AX pocket by Glide SP (colored by element). The Acetylated Lysine in purple color, a nearby Glu323 (in stick representation, colored by element), NAM is first docked into C pocket and is part of the receptor for NAD+.
RC (12-30): Please comment further on X pocket and its proximity to/overlap with peptide binding pocket. Does AX binding require peptide?
PL (12/31): The AX pose was obtained using SIRT3/AceCS2 from 4FVT and docked NAM (in C) together as receptor, and The ring structure of nicotinamide moiety is close to N terminal of AceCS2. but not in close to the side chain of Acetylated Lysine. Yes. AX pose do requires peptide. We also obtained several pose without the presence of peptide, and the favorable position seems to have nicotinamide moiety extend into the peptide binding pocket.
RC (12/31): Ok, the questions below regarding the docking (or lack thereof) to the AB pocket in Sir2 in presence of peptide are pertinent to the issue of whether the AX binding mode is analogous to the AB binding mode in Sir2, or whether these binding events occur under different conditions.
RC (1/9): In the absence of peptide, how do the MM-GBSA scores of the alternate binding modes of NAD in SIRT3 compare to the score of the AC pocket binding mode?
PL(01/09/14): There is no crystal structures for NAD+ in SIRT3, so there is no in-place MM-GBSA scores for NAD+ in SIRT3 in the absence of peptide. I do have MM-GBSA scores for NAD+ in SIRT3 (using different receptor) in the absence of peptide using Glide (SP or XP). And here are the results and they highly depend on which crystal structure is used as receptor :
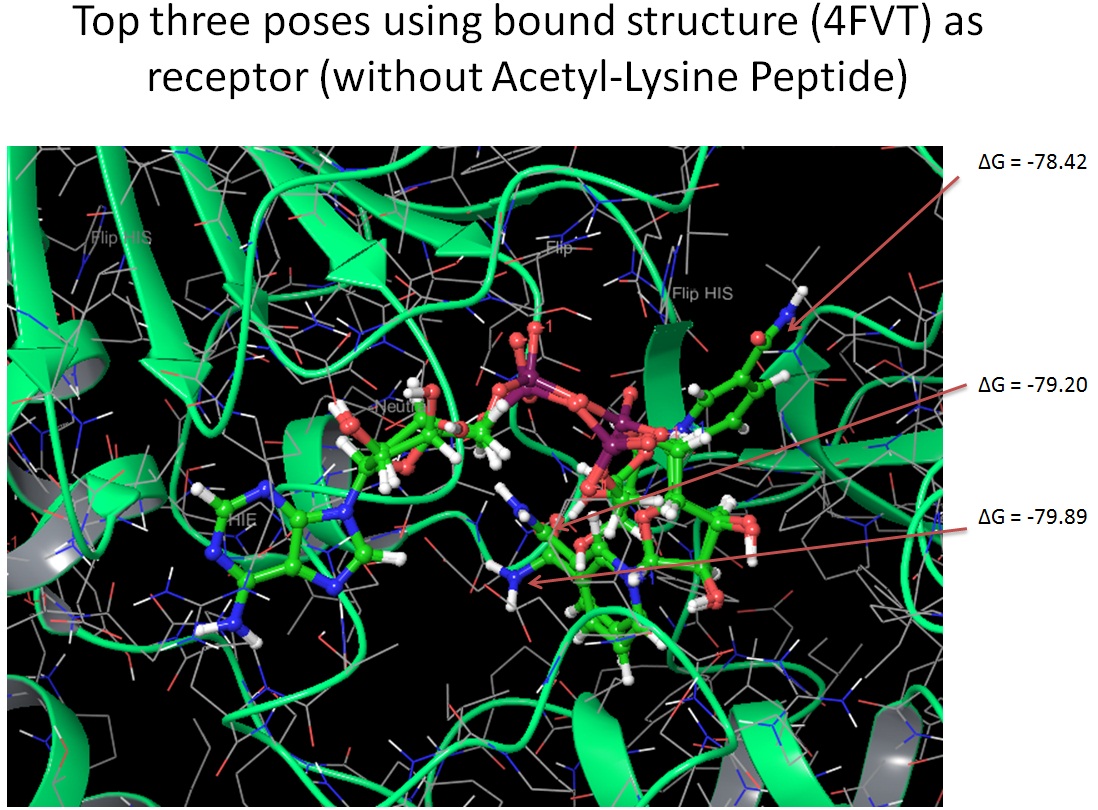
Using 4FVT as receptor, without peptide, NAD+ in AC pocket, -79.89, and we also observe a pose with nicotinamide moiety in the acetyllysine binding pocket with MM-GBSA score of -78.42.
Using 3GLS (apo-enzyme) as receptor, without peptide, NAD+ in AC pocket, MM-GBSA score is -67.72
Using 4BVG (intermediate step) as receptor, without peptide, nicotinamide moiety in acetyllysine binding pocket with MM-GBSA score of -100.88
Using 4JSR (enzyme with inhibitor) as receptor, without peptide, nicotinamide moiety in neither of the pocket mentioned above, MM-GBSA score is -86.29
Because we are using rigid receptor, the conformational change upon the binding of NAD+ only is not included.
For comparison, here is in-place NAD+ AC binding in SIRT3 (4FVT with AceCS2): -99.87
Please also comment on why the AX mode was not generated in the dockings using Eric's prepared structures.
PL(01/07/2014): Eric's docking for SIRT3 is mainly based on 3GLT (SIRT3 with intermediate), in which ARG158 was blocking the access to the B pocket. That combines with placement of other residues near B pocket may result in the failure to located AX binding mode.
n For the methods section revisions, we will need to describe MD methods used e.g., for the AX binding mode. Here is what I currently have from wiki postings. Please indicate if this is the version that can be used directly in the paper or revise/edit per the most recent simulations if needed.
"MD benchmark simulations are rather standard.
Amber 99SB force field is used.
For Zn, extra parameters are from the following reference:
1) Lu, Q., Tan, Y., Luo, R., Molecular Dynamics of p53 DNA Binding
Domain, J. Phys. Chem. 2007, 111:11538-11545
NAD+ was obtained from the following reference.
2) Walker, R.C, de Souza, M.M., Mercer, I.P., Gould, I.R., Klug, D.R.,
J. Phys. Chem. B., 2002, 106(44), 11658-11665
3) Pavelites, J.J., Gao, J.L., Bash, P.A., Mackerell, A.D., J. Comput.
Chem. 1997, 18, 221
Acetylated lysine was obtained from the following reference.
4) G.V. Papamokos,G. Tziatzos, D.G. Papageorgiou, S.D. Georgatos, A.S.
Politou, E. Kaxiras, Biophysical Journal 102 (2012) 1926-1933.
The initial structure of SIRT3/NAD+/peptide complex was constructed from 4FVT. Using AmberTools, extra 14 Sodium ions and 5 Chloride ions were added to neutralize the system, then the system was soaked in a box of TIP3P water molecules with a margin of 12 Angstrom. Prior to free MD simulations, a 10000-step minimization is performed with a constraint of 5 kcal/mol/A^2 on heavy atoms of complexes, followed by a 200 ps NPT MD under the same constraint. 2 fs time step was applied together with SHAKE algorithm on all bonds involving hydrogens. Langevin dynamics was used for temperature and pressure control (at 300 K). Electrostatic interactions were computed using Particle Mesh Ewald (PME) method. Simulations were carried out using NAMD on slave006 node (16 cores). Usually 2ns simulation can be obtained using 16 cores in 24 hours. MD simulation on SIRT3/NAD+/AceCS2/NAM complex has been carried out for 16 ns, and the system remain stable. Further MM-PB(GB)SA calculation using the obtained trajectory is underway. The RMSD of the protein backbone is attached here."
PL(12/27): Most of the above information can be used. Some extra details such as the use of slave006 node and run time can be omitted. Now longer simulation has achieved, and I can update sometime soon.
2) Experimental literature data:
n Please summarize the experimental binding affinity data that has been reported in the literature for NAD binding to Sir2,SIRT1 and SIRT3, with references. A range of values was reported for SIRT1. How did these experiments differ? (Either PL or XG)
XG(12-30): There are a variety of reports of NAD+ Km values for sirtuins. These measures establish that the enzyme affinity for NAD+ is not of high affinity and in the range of 15-560 mM for human enzymes. Will update if any refs were missed.
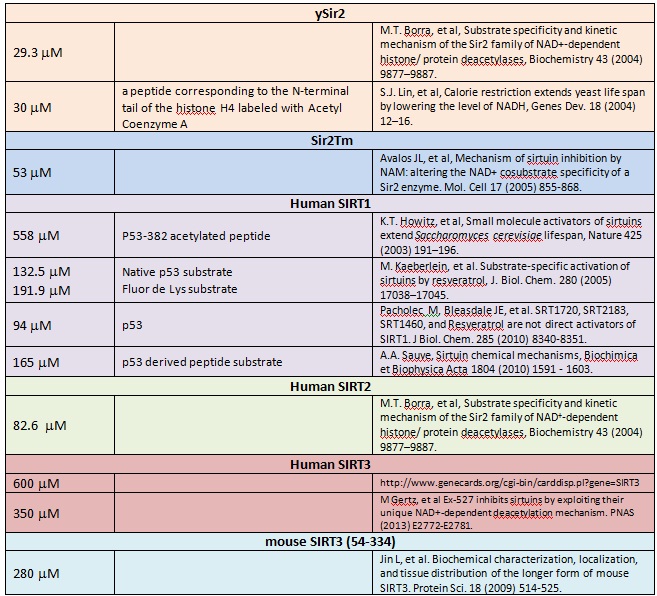
RC (1-10): Since the second column above refers to peptide substrate cocomplexed, were all these studies done with carba-NAD, if they were direct binding affinity measurements rather than kinetic estimates? And, please indicate whether any of the above studies were done w ITC.
Substrate specificity and kinetic mechanism_2004_Borra.pdfSubstrate-specific activation of sirtuins by resveratrol_2005_Kaeberlein.pdfSRT1720 SRT2183 SRT1460_2010_Pacholec.pdfSmall molecule activators of sirtuins extend SC lifespan_2003_Howitz.pdfMechanism of sirtuin inhibition by nicotinamide_2005_Avalos.pdfCalorie restriction extends yeast life span _2004_Lin.pdfSirtuin chemical mechanisms_2010_Sauve.pdfEx527 inhibits sirtuins by exploriting their unique NAD+dependent deacetylation mechanism_2013_Gertz.pdfBiochemical characterization, localization and tissue distributrion of the longer form of mSIRT3_2009_ Jin.pdf
n XG commented briefly on the relative concentrations of NAD, NAM, etc used to produce Wolberger's crystal structures containing NAM bound to the C pocket with NAD in the AB pocket. XG, please provide this information if it is available.
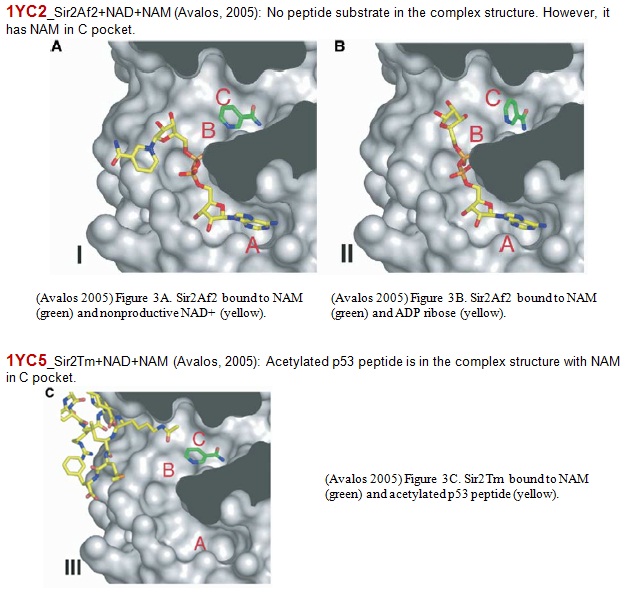
XG(12-30): Purified Sir2Af2 enzyme was dialyzed into 10 mM HEPES (pH=7.4) with 1 mM Tris (2-carboxyethyl)-phosphine TCEP and concentrated to 20 mg/ml. Prior to crystallization trials, 5.5 ul of a neutralized solution of 100 mM NAD+ was added to 50 ul of the Sir2AF2 solution to a final concentration of 10 mM NAD+. Crystals were grown by the hanging drop method in 0.1 M HEPES with 1.8 M ammonium sulfate, 1% PEG400, and 70 mM NAM and formed in space group P21212 with unit cell dimensions a=105.1Å, b=181.6Å, and c=79.0Å.
Purified Sir2Tm enzyme was dialyzed into 10 mM HEPES (pH=7.4) and concentrated to 16 mg/ml and 5ul of a 40 mM solution of acetylated p53 peptide (372-KKGQSTSRHK-K[Ac]-LMFKTEG-389) was added to a final concentration of 10 mg/ml Sir2Tm, and 4mM peptide. Crystals were grown by the hanging drop method in 100 mM CHES (pH=9.6) with 16% PEG3350, and 100 mM NAM and formed in space group P21212 with unit cell dimensions a=46.1Å, b=59.8Å, and c=106.2Å
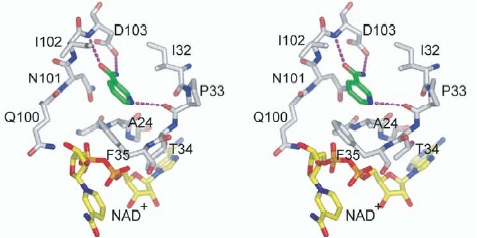
C pocket of Sir2Af2 (white) bound to nicotinamide (green) and nonproductive NAD+ (yellow).
n Wolberger presented a figure that showed multiple possible binding poses for NAD+ in the absence of peptide in Sir2 (these structures may have been reported earlier by Avalos 2004, Min 2001), and indicated that in the presence of peptide, NAD+ binds to the AC pocket. Please comment on whether any of the alternate binding poses of NAD+ mentioned by Wolberger overlap with the AX pocket in SIRT3 (if they are in the pdb). Do all these poses have NAM in the C pocket? Please also comment on whether or not AB pocket binding is observed computationally (docking) in the presence of peptide. Please comment on whether AC pocket binding is observed computationally (docking) in the absence of peptide.
Mechanism of sirtuin inhibition by nicotinamide.pdf
wolberger_sir2_structure_2004.pdf
RC (12/30): The above paper appears to indicate NAD binds only in the AC pocket in presence of peptide, in a strained conformation, and NAD binding to AB pocket is more favorable in absence of peptide. This information is important for our paper, so please have a look. Previously, we did not receive indication that NAD binds only to AC pocket in presence of peptide. This is related to the question above regarding whether NAD docking to the AB pocket is observed in computational studies in presence of peptide; this was not clear based on our earlier correspondence.
RC (1/09): PL, please see the questions in bold above and let us know whether docking of NAD to the AB pocket of Sir2 is observed in the presence of peptide. Whereas in the case of SIRT3, binding occurs to the AX pocket, where X is close to the B pocket, in the presence of peptide. Wolberger suggests that the AC pocket binding mode in Sir2 is a strained conformation compared to the AB pocket binding mode. Is there any evidence for this from docking or analysis of (e.g. in place scoring of) her structures?
After receiving this information, we will need to consider how to revise the section in the paper that compares the AB/AC and AX/AC pocket binding modes in Sir2/SIRT3. The data appears to suggest that NAD binding in Sir2 occurs to the AB pocket in the absence of peptide, and a conformational shift to the catalytically active AC pocket occurs upon peptide binding; whereas in SIRT3, binding occurs to the catalytically active AC pocket in the presence or absence of peptide. Some existing figures may need to be revised, and the SIRT3 AX pocket figure above will need to be referred to in this section. At this point we will also decide on whether to include any binding affinity experimental data or additional MD binding affinity calculations for NAD.
PL(01/09/14): In regard to the NAD binding in the presence of peptide, the top ranked pose is the NAD+ in AC pocket if 4FVT is used as receptor. However, adding NAM to the C pocket and we found AX binding mode, which seems to be confirmed by MD simulation up to now.
My previous calculations are all focus on SIRT3, and the only calculations using Sir2 are in-place MM-GBSA calculations using 1YC2, in which all the non-relevant molecules are removed, therefore no peptide is included.
I can carry out the docking using Sir2 structures that do include peptides and will report the output here. And the focus will be on whether we can find AB/AC binding mode in the presence of peptide.
I think there are strains on both AB and AC conformation, and maybe I can evaluate it using OPLS_2005, but I am not sure how accurate the result will be.
I just did a energy minimization on all non-polar hydrogens (cause least change to the structure) using prime (energies with VSGB solvation model), and here are the four energies for NAD+ at their poses in chain A-D.
NAD+ in 1YC2 chain A (AB mode 1) Prime energy w/ VSGB: -23.8
NAD+ in 1YC2 chain B (AC mode) Prime energy w/ VSGB: -22.7
NAD+ in 1YC2 chain A (AC mode) Prime energy w/ VSGB: -23.9
NAD+ in 1YC2 chain A (AB mode 2) Prime energy: -19.7
If change the setting to vacuum, here are the Prime energy obtained for each poses:
NAD+ in 1YC2 chain A (AB mode 1) Prime energy: 39.7
NAD+ in 1YC2 chain B (AC mode) Prime energy: 22.9
NAD+ in 1YC2 chain A (AC mode) Prime energy: 38.4
NAD+ in 1YC2 chain A (AB mode 2) Prime energy: 23.6
I am not sure if there is anything we can say about strains in the molecules from these numbers, and I need to check if the charge assignment is reasonable. If you think these calculations will be useful, I can check further.
RC: Yes, the docking to Sir2 in the presence of peptide is important. By constraints, are you referring to the strain in the AC mode referred to by Wolberger?
I haven't done any simulation on Sir2 so far, maybe I can carry out some of these calculations after docking in the presence of peptide so we can compare.
XG(12-30): Free NAM binding was observed in the C pocket in both complex structures of 1YC2 and 1YC5. 1YC2 (Archaeoglobus fulgides Sir2Af2): The crystal was grown in the presence of NAD+, PEG400, and NAM. The crystals contain five crystallographically independent monomers in the asymmetric unit that are in differently liganded states. Two of the five monomers (Chain A and D) in the asymmetric unit are ternary complexes containing NAM bound in the C pocket of the active site of Sir2Af2. One of these is also bound to NAD+ in a nonproductive conformation. Another ternary complex also contains alpha-ADP-ribose bound in the active site. A third complex (Chain E), which contains NAD+ bound in a nonproductive conformation, has density suggestive of NAM bound in the C pocket at low occupancy and is therefore no used in the paper. The remaining two monomers (Chain B and C) in the asymmetric unit are bound to NAD+ in a productive conformation that occupies the C pocket and to a PEG molecule that lies in the acetyl-lysine binding tunnel. There are two additional NAM molecules far from the active site that mediate apparently nonspecific interactions between monomers in the asymmetric unit.
1YC5 (Thermophilic bacterium Thermotoga maritima Sir2Tm) was in complex with NAM and an acetylated p53 peptide. A single well ordered nicotinamide molecule is bound to the C pocket of Sir2Tm.
Please also check Ping's posting above for details of the manual alignment between 1YC2 chain A (NAD+ in AB pocket, NAM in C pocket) and 4FVT with NAD+ docked into AX pocket by Glide SP.
A summary of Sirtuin structures up-to-date is attached. Summary of Sirtuin Structures 2013 update.docx
RC (12-30): It appears that Fig 1a in Wolberger 2005 (NAD in unproductive conformations in absence of peptide) is based on crystallographic structures; since they did not observe AC pocket binding in absence of peptide, they concluded AB pocket binding is more favorable. Fig 1b (NAD in AC pocket in presence of peptide) is based on the structure mentioned above with PEG in the peptide binding pocket; hence they concluded that peptide binding causes the NAD to bind only in AC pocket.
XG(12-31): The current observed crystallographical evidence shown that either peptide substrate or PEG type molecule (mimicking peptide binding) causes the NAD to bind only in AC pocket. When C pocket was occupied by other molecule like NAM, NAD binds to AB pocket in the absence of peptide binding. For 1ICI, Min et al propose that deacetylation takes place at B pocket. Later Wolberger's group indicated that AC pocket is NAD productive conformation. The examples are listed as following
RC (12-31): Ok, it will still be important to get the info regarding the docking results requested below.
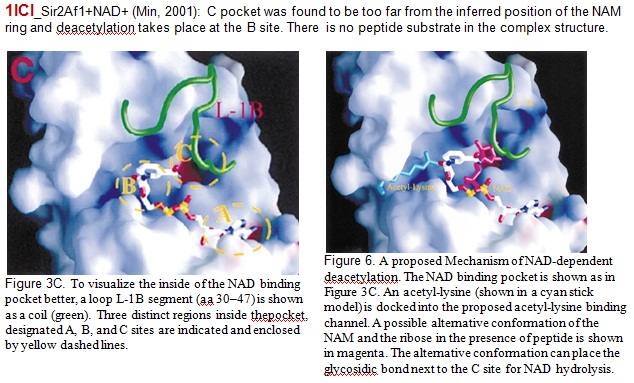
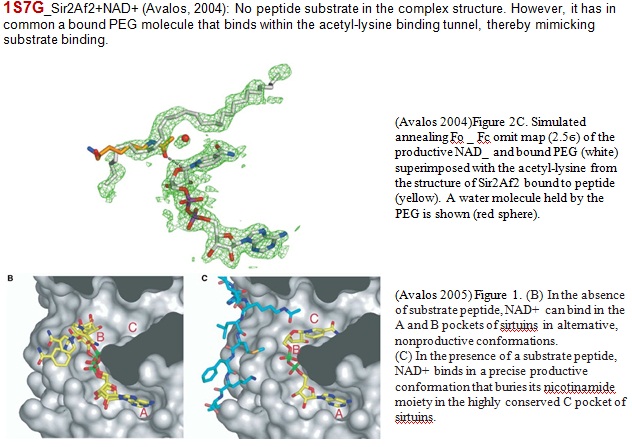
Our docking studies placed NAD in AC pocket or AB pocket in absence of peptide, but it was never observed crystallographically in AC pocket in absence of peptide. Please confirm. For SIRT3 (new figure above), we have AX binding in presence of peptide. Only in presence of peptide?
XG(1-07): Our in place docking studies from crystal structure 1YC2 were preformed in the absence of peptide. Yes, in the absence of peptide, NAD+ was not observed crystalographically in AC pocket. Actually, there was only one crystal structure (1ICI_Sir2Af1) reported so far without peptide. NAD was not observed in AC pocket, but not in AB pocket neither.
RC (1-09): In a figure (which I mentioned above) in her 2005 paper, Wolberger shows several different binding modes in/near the AB pocket in the absence of peptide. I assume there is only one in the pdb.
XG (1-09): In Wolberger Sir2 structure paper (2004) above, she mentioned several different structures (from two single crystals) in the absence of peptide. (1) A single crystal of Sir2Af2 grown in the presence of NAD+ and PEG 400 was reported with PDB ID: 1S7G. There is no peptide in these complexes structure. However, PEG400 was found within the acetyl-lysine binding tunnel. It indicated that PEG400 mimicked peptide binding (Figure 1 C in original paper). (2) In Figure 1B, the structure of the nonproductive NAD+ molecules bound to Sir2Af1 (PDB ID: 1ICI_Crystal structure of a SIR2 Homolog-NAD complex_2001_Min.pdfMin J., Landry, J., Sternglanz, R, and Vu, R.M. Crystal structure of a SIR2 homolog-NAD complex. Cell_2001_105: 269-279.) was reviewed. In these complexes, no peptide or PEG was in the crystal structure and NAD+ was observed in near AB pocket.
In Wolberger Mechanism of sirtuin inhibition by NAM paper (2005) above, she mentioned two structures (from a single crystal with PDB ID: 1YC2) in the absence of peptide. Figure 3A and 3B shown that no peptide substrate in the complex structure. However, C pocket was occupied by NAM.
RC (1-10): I was referring to #2 - Fig 1B, which refers to 1ICI,Sir2Af1. I now understand you mean that there are several nonproductive conformations that are in fact close to the AB pocket, though not all are exactly in it. To confirm, In 1YC2 with NAM in the C pocket and nothing in the peptide binding pocket, NAD is found in the AB pocket.
(to PL:) Do you confirm that NAD+ has not been observed in the AB pocket crystallographically, and if so, why was the AB pocket mentioned as a possible binding pocket for NAD in Sir2 (moreover, why has the B pocket been referred to as a pocket at all if nothing has been found to bind in it).
After double confirmed with Ping, Yes, for SIRT3 we have AX binding only in the presence of peptide. Ping’s docking studies of NAD in the presence of peptide for bound structure (4FVT) shown the lowest scale was in AC pocket.
Without peptide, scales were dropped.
RC (12/27): A list of data (including literature data) pertaining to the binding modes and affinities of NAD,NAM,isoNAM in Sir2/SIRT3 as they pertain to the inhibition mechanism is under preparation and will be posted shortly. Some of the data requested above will be included.
[More to be posted shortly]
XG (12/17): My current focus are
(1) Modifiy and revise the PCB manuscript for resubmission.
(2) Overnight purification to get rid of reduced glutathione. Then the protein will be concentrated. Hopefully the protein can be measured at the end of the week. It will be ready for the continuous assay.
(3) Discuss with Ping and have a list of small molecule candidates for Dr Raj to review. Because it will take 2-3 weeks to order and ship to the lab.
XG(12-16): The methods for base-exchange experiments is attached. It was partially posted here and there. I put them together. Methods for Base exchange.docx
PL & XG (12/16):
The update manuscript so far and responses to reviewers are attached.
Response to Reviewers' comments_12162013.docx
Mechanism of Inhibition_12152013_PL_XG.docx
The docking results from last week is also attached.
Sirtuin_Project01_Week7.docx
PL & XG (12/09):
This week's report included the continued effort in finding potential hits. And a selection of molecules will be finalized next week for your approval and followed experimental testing.
The docking results is attached. Sirtuin_Project01_Week6.docx
XG made changes to the manuscript according to reviewer's comment, and is attached here. Response to Reviewers' comments_12062013.docx
RC (12-13): I am having difficulty identifying where changes to the manuscript were made. Was XG planning to upload a revised paper draft here in addition to the above document? Please address those questions directly asked by the reviewers, most of which we discussed at our last 2 paper review meetings, at this time, and then we can have another paper review meeting to go over the revised draft. Please see my comments on priorities for revision vis-a-vis interpretation of the new computational results also posted to the Ping's tasks page. Thanks.
XG & I have been working on the kinetics model and the rate laws for each under rapid equilibrium approximation were presented in the response. And we are currently working on the rate equation using steady state approximation. Experimental data will be re-analyzed using the current models, and currently the fitting to the simplest model was achieved
RC: These derivations are somewhat analogous to the rapid equilibrium and quasi-steady state bi-reactants MM models Chaoran and I developed for the extension kinetics paper. It is good that we are working on them, as the development of these kinetic models was one of the objectives listed on the sirtuin overview page, and they may be needed for the 2nd paper. But it may be overkill to include much of this in the current paper, because: a) the paper has already been submitted and reviewed, and the referee only mentioned that the inhibition background included was well-known and should be reduced; b) supporting information is generally not read; c) the material would likely get better visibility in a separate work; d) fitting to multireactant models would change the results of the paper. e) Fitting to multireactant models can involve variation of the concentrations of multiple species in order to properly estimate all model parameters (e.g., to get the various Km's rather than just apparent Km's). We will have to discuss whether to include any of it in the current paper (if so, we might include the simplest model if there is space; we can discuss the fitting you are doing to that model after some of the other priority revisions are finished. This may be warranted if the results differ significantly from those currently presented, but we may always consider presenting the current results under the assumption of a standard competitive/noncompetitive framework). Our original plan was to treat this in a subsequent paper - either a primarily experimental one based on empirical model fittings, or the 2nd paper where we test various inhibitors in presence of NAM base exchange inhibition and compare model-obtained Ki's to computational binding affinity estimates. We should not proceed with quasi-steady state derivations at this time until we have laid out the plan, since they would certainly not be included in this paper.
A priority for now is the updating of the manuscript with the latest MM-GBSA data that seemed to resolve the issue of robustness of the scores with respect to the choice of crystal structure, according to the plan recently indicated on the wiki. There I asked whether we are now able to address most of referee 2's questions, and whether the only remaining simulation required is the MD for the AB pocket. If so, please complete all those outlined changes except for the MD in AB pocket.
MD simulation on SIRT3/NAD+/AceCS2/NAM complex has been carried out for 16 ns, and the system remain stable. Further MM-PB(GB)SA calculation using the obtained trajectory is underway.
The RMSD of the protein backbone is attached here.
Here is a snapshot from the simulation, NAD+ (colored based on atom type), Acetylated Lysine (pink), and NAM (green) are in ball-and-stick representation.
RC (12-13): Please let me know if the MD simulation of SIRT3 NAD+/NAM above is the one discussed with NAD+ placed in the AB pocket via a superposition of the Sir2 AB pocket structure
PL (12/13): Yes. The simulation above started from placing NAD+ in the AB pocket.
PL & XG (12-2): progress report is attached
Progress report 12.02.2013.docx
PL & XG (11-25): progress report is attached
Progress report 11.25.2013.docx
RC (11-25): Thanks. What is the next step for computation? Are we bottlenecked by the need for experimental screening at this time, or are we continuing to screen larger library sizes? When do we plan to shortlist the molecules for experimental screening, and how many? At what point would the computational part of project 1 shift more focus to LIA or MD?
PL(11/26): From the current screening results, I will be suggesting some molecules for experimental tests. Based on the characters of molecules that do not fit or produce low MM-GBSA scores, I have been filtering out some molecules from the current molecular library. Even though we are targeting small molecules, (m.w. < 200), there is no way we can cover all the drug-like molecules due to the limitation of the license and computing power. However, I do suggest that we explore as many molecules as possible, which will increase our chance of finding some good leads, and produce enough amount of work for a better publication.
RC (11/28): Please give me an idea of how many more molecules we would like to dock before experimental screening, and whether there are any other licensing options we should be looking at. For example, I seem to recall licenses that allow for more instances of glide to be running concurrently on a cluster. We are not exploiting our batch server at all at this time. Or would the time required for structure preparation render this useless? I am currently communicating with Schrodinger about pricing.
PL(12/4): Actually, I have posted the license information on 10/30 on this page. We currently have token-based license, which give us access to most of the tools in Schrodinger's suite. And with 25 tokens we currently have, we are limited to 4 Glide jobs or 3 Prime (MM-GBSA) jobs to run concurrently. Under the token-based license, the only way is to get more tokens in order to increase screening throughput. We can change the licensing scheme, and request extra licenses specific to Glide and Prime modules and reduce the number of flexible tokens (8 minimum). You have to weigh on the pricing and what the future use of Schrodinger's suite. One potential use is the QSite module for QM/MM calculation, although I will probably prefer other codes to do it.
Currently, the bottleneck of our screening workflow is on the Glide and Prime calculations. Small molecule preparation and selection doesn't take as much time. I have already prepared over 3000 small molecules and over 10,000 isomers.
At the same time, I have established the whole protocol of generating force field parameters for selected molecules, e.g. using firefly to carry out structure optimization using QM method, then derive the charges for the new molecules using RESP method, other force field parameters will be obtained from GAFF, and the topology will be built using antechamber.
No MD has been set up for these docked results. And we have to set priority between the PCB project and the current project so we can manage the time and resource better.
RC (11/28): It seems some of the issues with time management will be mitigated now that we have gotten better results with single structure MM-GBSA for the PCB paper.
RC (12-26): Regarding licenses: Since one of our focuses with virtual screening is the discovery of small molecules that bind in the C pocket, we should benchmark the free docking software in terms of its ability to generate poses for small molecules that bind in that pocket (e.g. NAM). Given that C pocket binders may have a relatively small number of degrees of freedom, is Glide necessary? Please comment on pose sampling vs scoring function advantages of Glide in this context.
PL & XG (11-18): progress report is attached.
Progress report 11.18.2013.docx
RC (11-19): Are we on track for completion of the enzyme preps for the continuous assay by Dec 1? Are we simply obtaining more enzyme for the assay at this time? If so, how did we settleon the amount of enzyme to prepare?
XG(11-19): In the process of enzyme preps, we have gained the following strength: (1) a certain amount of protein; (2) a solid and fast protocol to prepare more proteins because you will not make enough enzyme by one or two batches for future work. Talking about how much is enough, the amount of enzyme we need for assay is depent on the activity of enzyme and how many inhibitors we are going to test. The enzyme concentration and activity of every batch may vary. The next step is characterization of enzyme and test for continuous assay. The enzyme prepration will be carried on whenever more enzyme is needed. On the side, I am working on responding reviewers' question for PCB manuscript. It needs continuous efforts. I may have to stop lab work for sometime to finish revising PCB manuscript.
RC: How long do you think enzyme characterization and continuous assay test will take? I assume we have not done these steps before.
XG(11-19): 1 week for enzyme characterization and ~ 2 weeks for continuous assay test.
RC: Are these the immediate next steps?
XG(11-20): Yes.
PL & XG (11-11): progress report is attached.
Progress report 11.11.2013.docx
PL(11/05): Report for week 1 is attached.
Sirtuin_Project01_Week1.docx
XG (11-4): Optimize purification purcedure. Small scale purification was performed. The GST-tagged pGEX-6P3 - PNCA protein was successfully purified. Some loss of protein were detected at the flow through step. To improve the yeild. FT step need furture modification.
PL/XG(10-25), Detailed plan is posted.Detailed-Plan for project 2-sub1.docx
Note: It was found that, for sirtuin paper, the articles published at PNAS/Molecular Cell/Structure/JACS most likely contain a portion of cocrystal structure work. For the articles published at Nature/JBC contain a portion of in vivo data. RC: Agreed, so it is important to have some feedback from the CROs on crystallography(; this is particularly important for the classes of inhibitors which induce larger conformational changes in the receptor).
RC(10-25): Thanks. Please see my comments/questions attached. We can arrange to meet to discuss later today (please let me know your convenient time). PL will also be preparing more details on the projects for other classes of inhibitors. I have asked him to start to prepare more detailed workflows/statements of novelty for perhaps two other classes; these can start with an outline of each, followed by development of documents akin to what you have posted for this project. The computational/experimental workflows may differ for these projects due to different amounts/types of computational sampling required, the achievable accuracy of binding affinity estimates, etc. We can then allot appropriate amounts of time in the schedule for one of these secondary projects.
Detailed-Plan for project 2-sub1_RC.docx
XG(10/25): Minutes on 10-25-13 afternoon meeting (RC/PL/XG)
(1) Went through the proposal “Plan for project 2 – subproject 1: Activator design targeting base-exchange reaction”.
Please check the attached file for details. Detailed-Plan for project 2-sub1_response to RC.docx
(2) RC: When we prepare our paper, we need to compare previous work, such as “Thiobarbiturates as Sirtuin inhibitors: virtual screening, Free-energy calculations, and biological testing” Thiobarbiturates as Sirtuin Inhibitors Virtual Screening Free-Energy Calculations and Biological Testing (2).pdf, to emphasize the significance of our work, IC50 falls in uM range but high specificity…
(3) RC: On the practice of pharmaceutical applications, one may ask if tight binding is the only way to measure if the drug is good or not. Practically, there are other factors need to be taken into consideration…
(4) RC asked for experimental approach of measure protein-ligand binding affinity.
PL: PNAS paper shows that the use of microscale Thermophoresis. It provides not only binding affinity but also binding mode.
XG: Please check the attached pdf for detailed application of Thermophoresis. Thermophore.pdf
XG need to start requesting for quote.
(5) PL will provide a proposal for 2nd project. XG will cooperate with experimental details in the near future.
RC (10-29): Please provide a rough weekly schedule for the computational activator discovery work over the next month or more (including when you will do LIA and MD if needed). MD will be essential for phase 2 (lead optimization) anyway; we may then need to augment binding affinity calculations w on-off rate calculations for which accurate binding affinity prediction will be a prerequisite. You should allocate about 1/4 of your time to proposed workflow for other classes of inhibitor discovery; that time allocation may soon (within next 1-2 weeks) be replaced in part by tasks for PCR project, which RC will comment on shortly.
PL (10/30): Computational activator discovery:
Week 1: dock 16 prepared ligands (as on detailed-plan) into 4BVG (enzyme only by removing intermediate) using Glide XP, followed by MM-GBSA evaluation. (necessary to correlate with their binding affinity alone); There is no special reason why to choose 4BVG instead of 3GLS (apo-enzyme) or other complex structures. However, the binding pockets in 4BVG are open and not biased as in some other complex (i.e. 4FVT or 4JSR, in which C pocket is occupied by NAM and ELT inhibitor in the complex structures.)
dock 16 prepared ligands (as on detailed-plan) into 4BVG with Intermediate using Glide XP, followed by MM-GBSA evaluation. (necessary to check there ability to inhibit base-exchange reaction)
search emolecule.com, hit2lead.com for a collection of commercially available small molecules (m.w < 200) for docking and MM-GBSA evaluation.
RC (11-4): Please provide an update here when each week's work is finished.
Week 2-4: dock the selected small molecules (~400) into the 4BVG (with and without intermediate)
Beginning week 2: set up MD on the cluster and perform benchmark and test on the system. set up software to generate force field parameters and topology fiels for selected ligands, non-standard residue, (Acetylated Lysine) etc.
After week 4: Provide XG with a few selections (with different binding affinity than already known values) for the test of IC50. find correlation.
One the next month of computations is nearly finished for phase 1, I would like a similarly detailed schedule for MD/phase 2.
RC (10-29): Please indicate whether use of the batch server would increase throughput of computational work and other details of how you plan to set up screening of 100 molecules in the schedule. Please confirm explicit water placement is not important for this project. For other projects, indicate how it will be done (e.g. using what tool - watermap or otherwise) where necessary.
PL (10/30): The limitation is mainly on the number of tocken, currently we have 25 token, which can support up to 4 Glide XP job (6 token each), 3 Prime job (eight token each). single computer with more 4 cores is able to use up all the tokens, therefore the server doesn't help the throughput. The location of explicit water around the C pocket highly depends on whether there is a bound ligand, and what ligand is bound. Comparing several complex structures suggests that explicit water can not be pre-determined. Explicit water away from binding pocket expect to have much less impact on the binding, therefore, not including explicit water is actually more consistent choice at this moment. Watermap is currently only available in commercial license, not included in academic license. It can be introduced in MD.
RC (10-29): You both indicated that SIRT3 likely undergoes a similar level of base exchange inhibition as SIRT1- please review the evidence for this (either PL or XG). Since isoNAM activation is much more pronounced for SIRT1, this suggests we may be able to develop isoform-specific activator leads (possibly as a theme of paper) and should screen all against SIRT1/Sir2 after we finish w SIRT3.
XG(10-30): All studies of NAM-exchange were carried on bacterial, mouse, and yeast Sir2. Among them, yeast Sir2 was used to study the activation effect by relief of NAM inhibition. Yes, we may be able to develop isoform-specific activator leads for SIRT1/Sir2 after SIRT3. It will be interesting if we can find some enzyme-specific activators.
The evidence that SIRT3 likely undergoes a similar level of NAM-exchange inhibition as Sir2:
(1) Current available IC50 values for ySIRT2, SIRT1, and SIRT3 are listed as following
| Reported |
PMC-AT lab |
|
| hSIRT1 |
~50 uM |
68.1uM |
| hSIRT3 |
~50 uM |
36.7uM |
| ySir2 |
100 uM |
XG (10-30): Reported physilogyical [NAM] are range from 10-150 uM. More specific
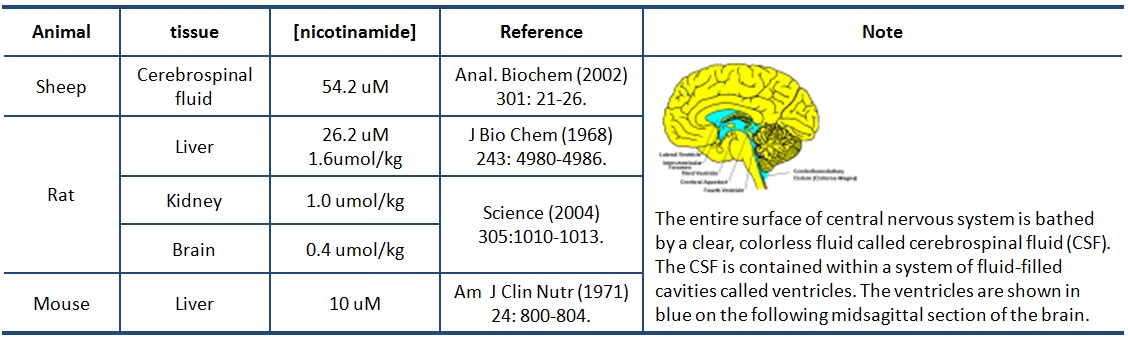
IC 50 shows that NAM is a potent inhibitor for ySir2, SIRT1, and SIRT3. This information is important because if NAM inhibition is not that significant, it will not be practical to design such type activator by NAM inhibition relieve.
(2) Sauve’s results show that nicotinamide did not cause complete inhibition for the bacterial and yeast Sir2 (% inhibition are 35% and 79%, respectively). For mouse Sir2, >95% inhibition occurred at high NAM concentration (1.2 mM). >95% inhibition was observed for SIRT1 and SIRT3 when NAM concentration is around 2mM.
(3) Sauve’s data: In the presence of 125 uM NAM, the ySir2 deacetylation activity was increased up to 50% by increasing isoNAM concentration up to 100 mM (Figure below).
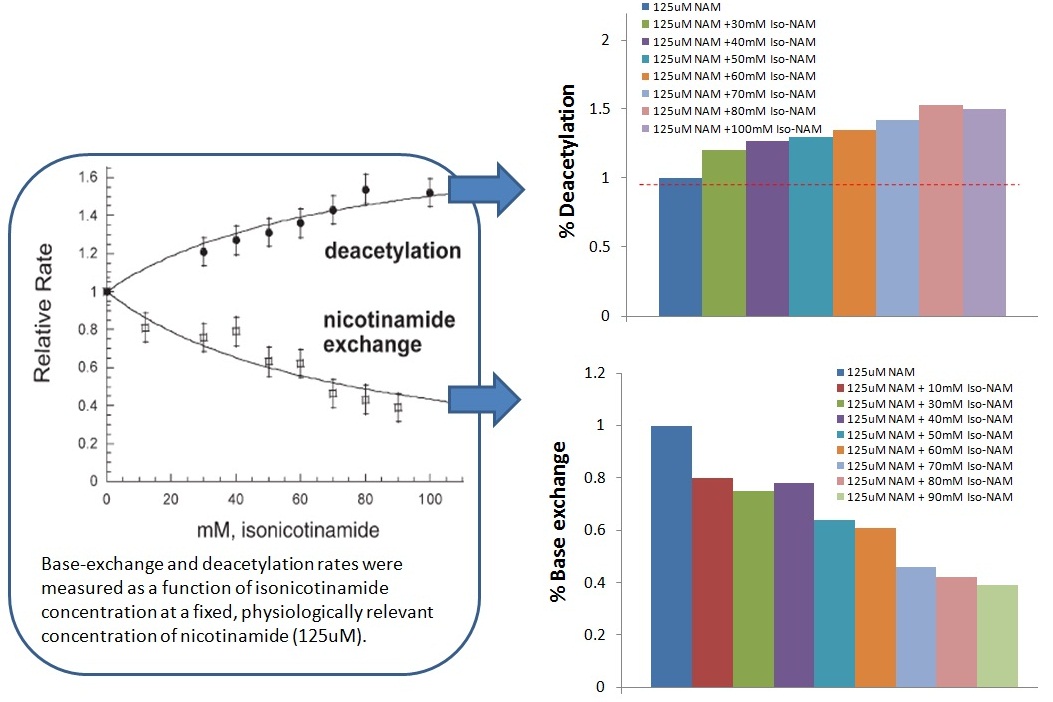
RC (10-30): To confirm, Sauve had no way of experimentally measuring the on/off rates of NAM or isoNAM?
XG (10-30): No.
(4) PMC-AT lab results: In the presence of 100 uM NAM, the SIRT3 deacetylation activity was increased up to 7% by increasing isoNAM concentration up to 900 uM.
(5) They are comparable (Figure below)
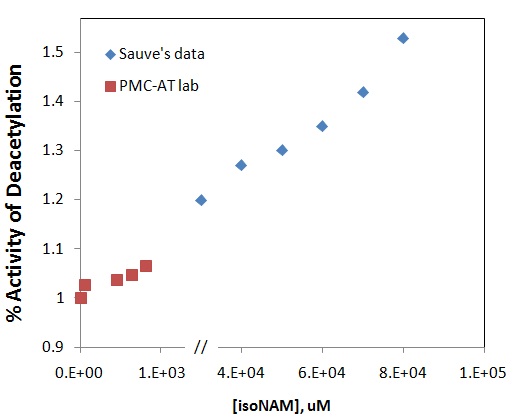
RC (10-30): Interesting - I didn't realize they were comparable; I only recalled modest activation for SIRT3. I suppose Sauve experimented with higher [isoNAM] than you.
XG (10-30): The [isoNAM] Sauve used up to100 mM and up to 900 uM of isoNAM was used in our experiments. Our results fell in the early stage of Sauve's curve. Please notice that there is a break in x-axis which indicates the concentration jump.
RC (10-30): There are two important points to bear in mind regarding the implications of these data for activator design (we discussed some of these before, but I'm repeating since they are relevant to this project):
a) Maximal possible extent of activation: It appears that physiological [NAM] is around the IC50. This appears to suggest that the maximal possible extent of activation in the body by alleviation of base exchange inhibition is around 2x? I looked at resveratrol for comparison. In the absence of NAM inhibition, sufficiently high concentrations lead to > 10x activation. Other SRT compounds may have substantially higher activation. In the presence of physiological [NAM], the extent of activation can be lower. We should bear in mind that our mechanism-based activators could be applied together with allosteric STACs like resveratrol since they would likely further increase the activation by STACs in the presence of physiological [NAM] - so we are not directly competing with allosteric activators like resveratrol. If/when in the future structural information on STAC-sirtuin complexes becomes available, any activators we identify can serve as leads for drugs that work in the presence of such allosteric STACs and lead optimization could be carried out using methods we are developing but starting with a STAC-sirtuin complex crystal structure.
If, after our initial screening, we are unable to find better activators than isoNAM, we can focus the presentation for this paper on accurate prediction of binding affinities of these small molecules, and then indicate that such a predictive model can be used to identify additional activator leads. We could then continue with phase 2 of this project, studying the mechanism of base exchange and its alleviation by isoNAM both experimentally and computationally (MD), since that understanding may facilitate the discovery of new activators. If this takes time and is deemed important, while doing so, we could continue with lead discovery of inhibitors that follow the other inhibitory mechanisms.
b) Drug concentration required: For physiological experiments with resveratrol, it seems people are using < 10 uM (2x activation is achieved around 10 uM). Given that much higher concentrations of isoNAM are required to activate sirtuins, one reason that these molecules are not being recommended for their health effects through sirtuin activation may be that they are not sufficiently potent. This is an opportunity and motivation for our project - to find activators that can alleviate base exchange inhibition at much lower concentrations. One way of examining the difficulty of this problem is to study the on/off rate constants of a potential activator that are required to achieve a specific level of activation at sufficiently low concentrations of activator, and to compare these rate constants to those of isoNAM. At this time, we don't have those on-off rates, but we can estimate them in phase 2 through MD. If there is any way to get them experimentally, we should be aware of it.
RC (11-1): More on base exchange activators vis-a-vis allosteric activators:
a) XG will look into the % activity enhancement of SIRT1 by resveratrol in the presence of 50 uM NAM and report to us.Is reservatrol activation substantially decreased by physiological [NAM] (i.e., is base exchange inhibition of SIRT1 increased by resveratrol)? If so, we are interested in understanding the mechanism of this, since it may then be possible to further boost activation by derepression of base exchange inhibition, using the rational activator design workflows we are developing in this project. One way is to measure the Km of [NAM] for SIRT1 in the presence of resveratrol. Nothing has been reported here. Do allosteric activators increase the binding affinity of NAM as well as peptide/NAD+? It appears this would be a straightforward experiment. We may plan for it when we have time in our schedule.
XG (11-1): Based on Sinclair's 2003 paper, the relative activity is shown below. In the presence of 50 uM NAM (close to physiological concentration), 5 uM of Resveratrol does not change activity a lot. Addition of 20 and 100 uM of Resveratrol increase Sir2 activity 3.5 and 3.6 fold.
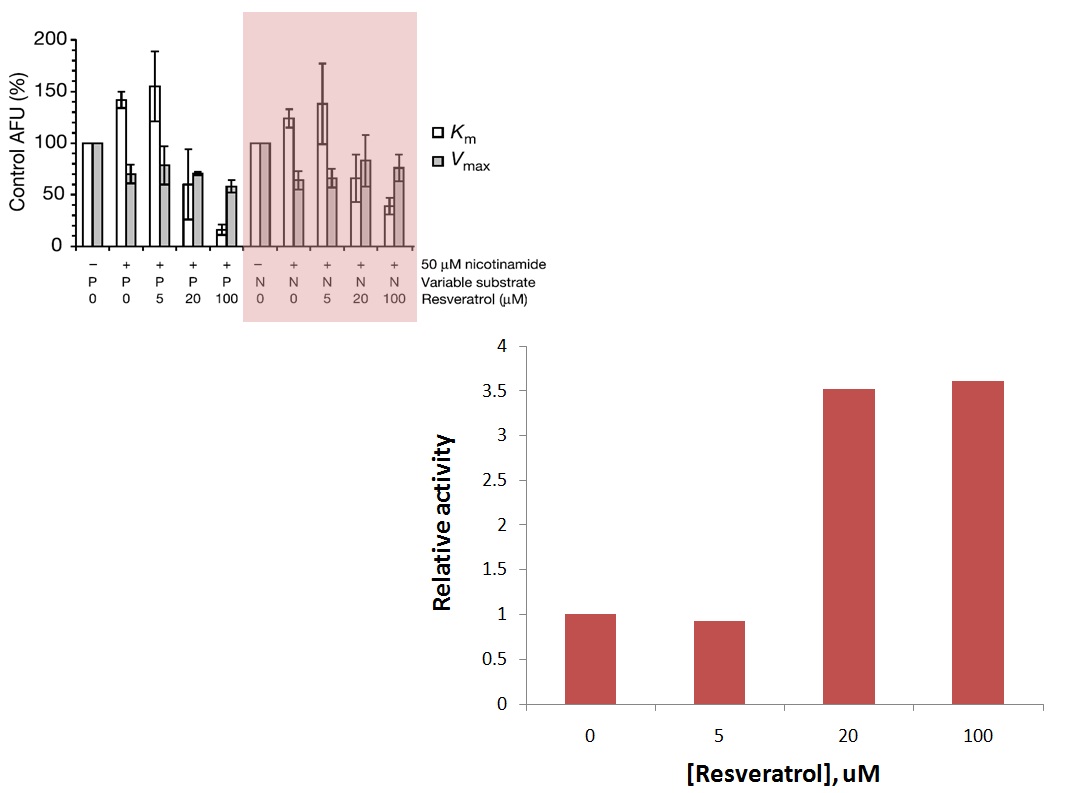
RC: Interesting, it appears that NAM then causes saturation of resveratrol activation at a lower fold enhancement. We need to explore whether this could be caused by the effect of resveratrol on NAM Kd. In the paper they claimed to be looking into these effects, but they were never published.
RC (11-2): Please also do a patent search on the idea of applying allosteric activators together with base exchange activators to boost sirtuin activity in vivo, and post what you find (if anything).
XG (11-12): Using SureChem database from Macmillan Publishers and Sirtris website, 566 patents were found. Lists of and Sirtris Pharmaceuticals, Lnc Patent applications were saved as pdf files. So far, no evidence was shown that "the idea of applying allosteric activators together with base exchange activators to boost sirtuin activity" has been patent.
recent patent applictions related to sirtuin modulators.pdf patents_assignee_sirtris-pharmaceuticals_2008-201.pdf
c) A 2012 paper solved the structure of SIRT5/reservatrol and claimed that there was a similar level of activation as in the case of SIRT1. However, binding occurred to the peptide pocket and activation was apparently caused by induced conformational changes in loops there. This does not appear to be the same allosteric mechanism as in the case of SIRT1 which apparently involved N-terminal resveratrol binding. Also, binding may have been facilitated by a fluorophore. What is the connection if any between this binding/activation mechanism and that observed for SIRT1? We remain skeptical regarding the use of this crystal structure to explain allosteric activation, but we should keep abreast of published crystal structures of allosteric activator complexes due to their possible use in computational modeling of effects like those described in a) above, and should consider the use of CROs for crystallography of our own lead complexes.
XG (11-5): Please check the attached file for details. Comparison between SIRT1 2013 paper and SIRT5 2012 paper.docx
RC (10-30): XG, you mentioned that the assay would take 1 month to finish (by which I assume we mean having enough enzyme to run a certain number of assays). If it is not finished by Dec 1, please start working on the screening of leads according to the experimental part of the protocol we have planned out (which I assume you could do with the continuous assay using whatever enzyme you have to date), and come back to make more enzyme later.
XG(10-30): OK.